
Featured research
ES&T | ExposomeX: Development of An Integrative Exposomics Platform to Expedite Discovery of “Exposure-Biology-Disease” Nexus
Manuscript Type: Full Article
First Author: Bin Wang (Peking University)
Corresponding Author: Mingliang Fang (Fudan University)
Journal: Environmental Science & Technology, IF2024 =11.3
Article info: Bin Wang, Changxin Lan, Guohuan Zhang, Mengyuan Ren, Tianxiang Wu, Ning Gao, Weinan Lin, Yanqiu Feng, Han Zhang, Bahabake Jiangtulu, Yuting Wang, Shu Su, Zhijian Liu, Xuqiang Shao, Fanrong Zhao, Bo Peng, Xiaotong Ji, Xiaojia Chen, Min Nian, Junjie Yang, and Mingliang Fang*. ExposomeX: Development of an Integrative Exposomic Platform to Expedite Discovery of the “Exposure−Biology−Disease” Nexus. Environmental Science & Technology, 2025, doi.org/10.1021/acs.est.5c05956
Full Text Link: https://pubs.acs.org/doi/10.1021/acs.est.5c05956

This article highlights the critical role of environmental exposures in human health and the growing importance of the exposome as a next-generation strategy for linking environmental factors to disease outcomes. Despite its conceptual appeal, exposome research faces major challenges that limit its advancement. First, integrating high-dimensional data—including external exposures, internal biomarkers, and disease outcomes—is highly complex, and the lack of standardized statistical workflows for different epidemiological study designs hampers reproducibility and accessibility, especially for new researchers. Second, uncovering biological mechanisms that link exposures to diseases remains difficult due to the absence of platforms that connect exposomic data with curated biological databases, although existing resources like CTD and STITCH demonstrate the feasibility of such integration. Third, mass spectrometry-based non-targeted analysis (NTA) offers broad coverage for exposomic profiling, but identifying disease-relevant features requires advanced statistical methods beyond traditional approaches used in metabolomics. Moreover, different analytical methods can yield divergent results, emphasizing the need for cross-method comparisons. Overall, an integrated, user-friendly platform that addresses these challenges is urgently needed to advance exposome-based environmental health research.
Platform Design
The integrated platform ExposomeX was developed with the aim of bridging the "Exposure–Biology–Disease" nexus (Figure 1). As the first comprehensive analytical platform for exposomic study, ExposomeX includes both an online web-based user interface (www.exposomex.cn) (Figure 1A) and an R package (https://github.com/ExposomeX). The platform comprised six major functions: statistical analysis (E-STAT), exposome database (E-DB), mass spectrometry data processing (E-MS), meta-analysis (E-META), biological link (E-BIO), and data visualization (E-VIZ). These functions are interconnected as follows (Figure 1B): E-DB supports E-MS, E-BIO, E-META, and E-STAT by providing basic information, e.g., environmental exposures, chemical toxicity, interaction network, statistical models, and effect size associated with various diseases; E-MS provides annotated features for E-STAT to build statistical models, and supports bioinformatics analyses by identifying key exposomic features, such as identified by NTA, for biological explanations; E-STAT provides the common statistical tools applicable to exposomic studies, enabling the generation of inputs for E-MS and E-BIO to elucidate mechanisms or validate novel findings; E-BIO offers potential biological links for selecting appropriate statistical models in E-STAT and E-MS with the aim of establishing an “Exposure-Biology-Disease” nexus; E-META enhances statistical interpretation by providing meta-analysis results for E-BIO and E-STAT; E-VIZ provides a visualization for exploratory analyses, using the results from E-META, E-BIO, E-MS, and E-STAT. These six functions were supported by 15 developed modules, including 10 for E-STAT (StatTidy, StatDesc, StatCros, StatMO, StatPanel, StatSurv, StatMedt, StatMix, StatHEP, StatIP), EDb for E-DB, EMS for E-MS, EMeta for E-META, EBio for E-BIO, and EViz for E-VIZ. Introductory videos are shared on the YouTube and Bilibili websites (Figure 1C).The related operation instruction materials and databases can be downloaded from the website: www.exposomex.cn/#/download (Figure 1D).
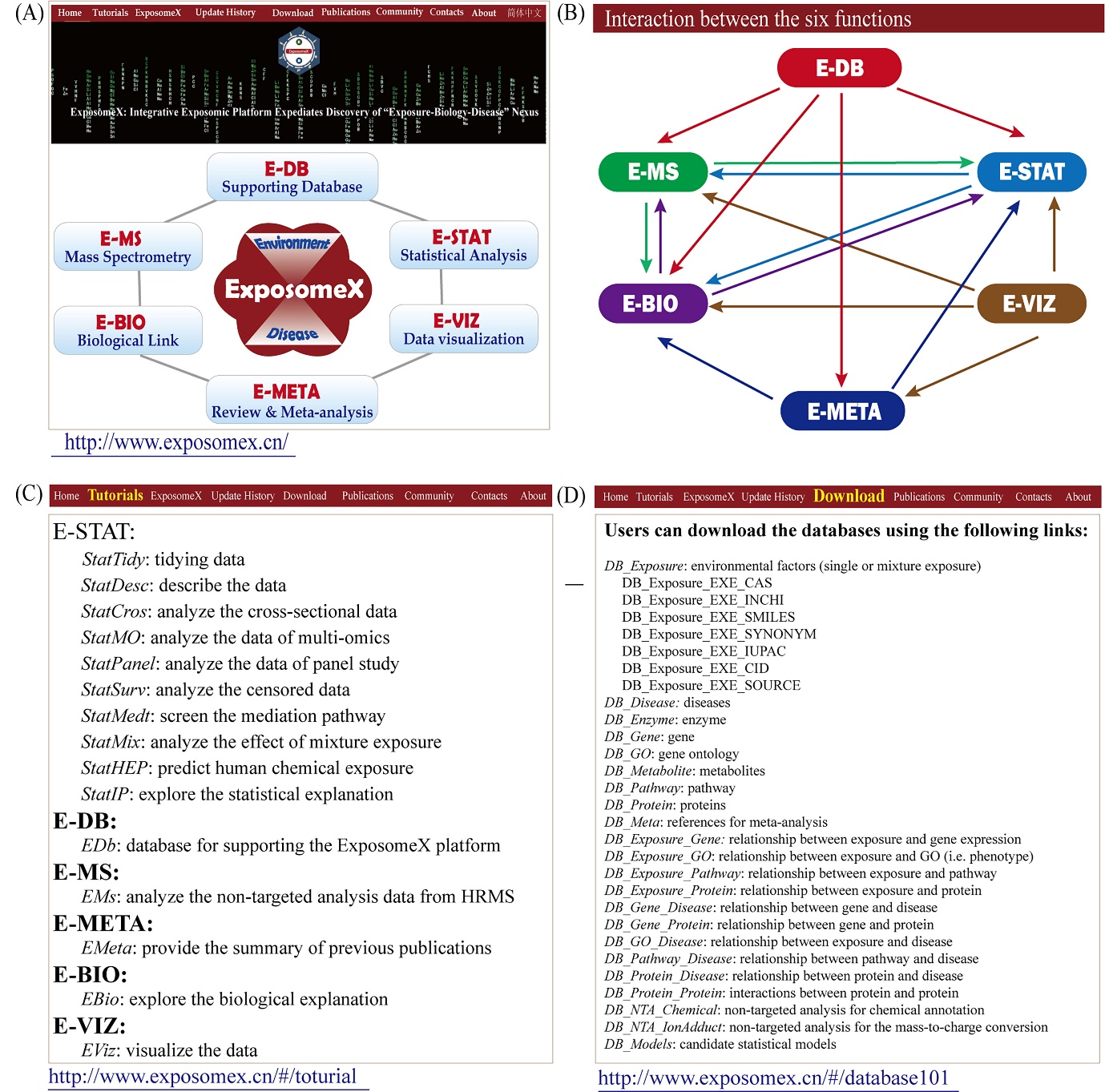
Figure 1. The ExposomeX platform design. (A) Main page of the web-based user interface; (B) Interactions between the six functions of the platform. The arrows indicate the supporting roles from the start function to the end function, with different colours distinguishing the connections of the individual functions; (C) User interface of the video tutorials for using the 15 supporting modules of the six functions; and (D) User interface for downloading the 23 curated supporting databases.
E-DB: Curation and Its Usage
The development of ExposomeX’s six core functions was supported by curated databases covering chemical exposures, biomarkers, protein targets, disease associations, and biological pathways. Data were integrated from established sources such as CTD, STITCH, ToxCast, and T3DB, with additional manual curation for non-chemical exposures like air pollution, diet, and lifestyle factors. The platform includes over 119 million unique exposures, 17,186 diseases, 153,780 proteins, and extensive gene, pathway, and GO annotations. Key exposure–biological interaction links, totaling over 100 million exposure–gene and exposure–disease pairs, were compiled for integrated analysis, as summarized in Table 1 and Figure 2A.
Table 1. Key Features and Data Sources of Databases Integrated in the ExposomeX Platform
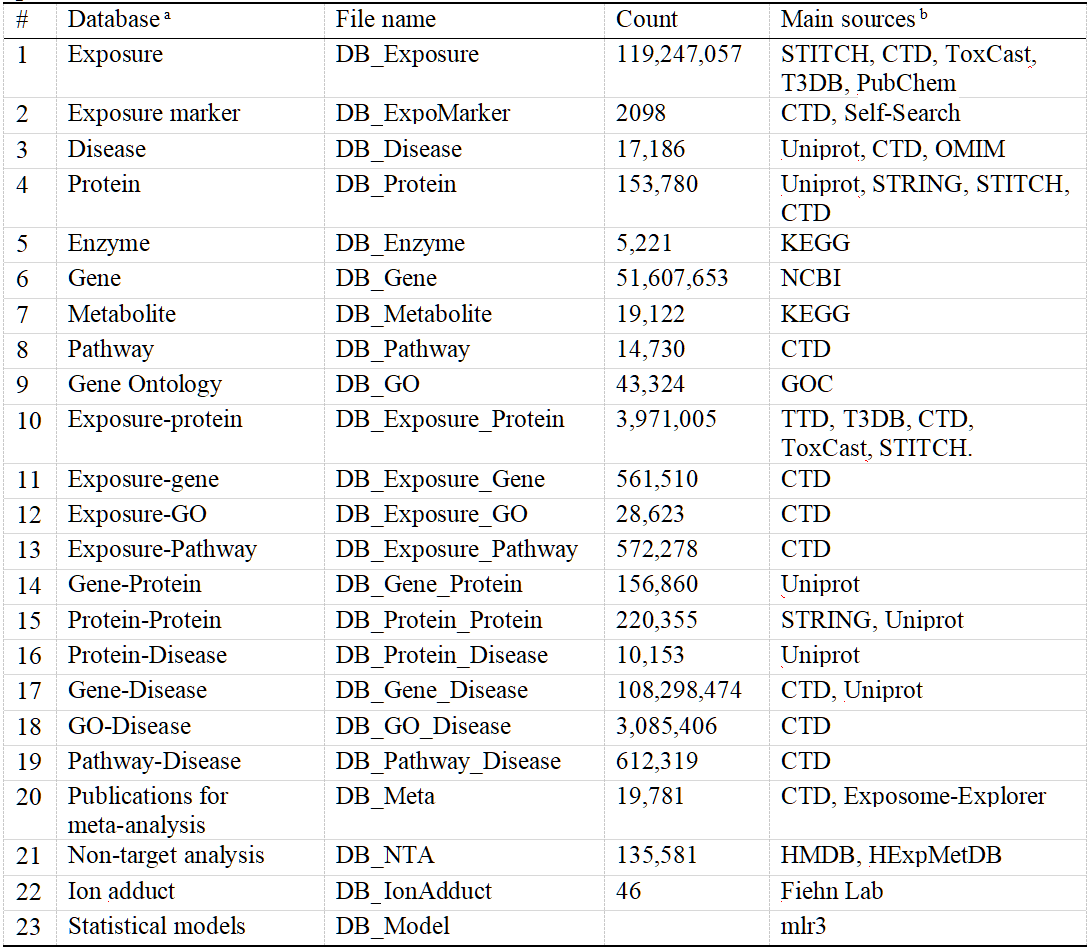
a Dataset download website (http://www.exposomex.cn/#/download);
E-META and E-BIO: Comprehensive Exploration of the “Exposure–Biology–Diseases” Nexus
In the current version of the E-BIO function, ExposomeX proposes several typical interaction modes to integrate the “Exposure-Biology-Disease” nexus, including EGoD, EGeD, EPaD, EPD, and EPPD (Figure 2B). For instance, EPPD was used to explore the network relationships between 13 environmental exposures and six diseases (Figure 2C), revealing potential shared protein targets and suggesting mechanisms of action and exposure-exposure interactions. Notably, diazinon and 2,3,7,8-TCDD emerged as key toxicants with mechanistic pathways like “Diazinon~EFNA2~KIT~Testicular germ cell tumor” and “2,3,7,8-TCDD~PLAT~IL-6~Rheumatoid arthritis”. The platform also supports literature-based synthesis via the MetaRef and MetaRev functions (Figure 2D-E), such as evaluating zinc intake and asthma risk. A meta-analysis (MetaAsso) on copper exposure and polycystic ovary syndrome, integrating 12 studies, provided forest plots and summary estimates using both fixed-effect and random-effect models (Figure 2F). Users can contribute new studies interactively through the platform’s community interface: http://www.exposomex.cn/#/community.
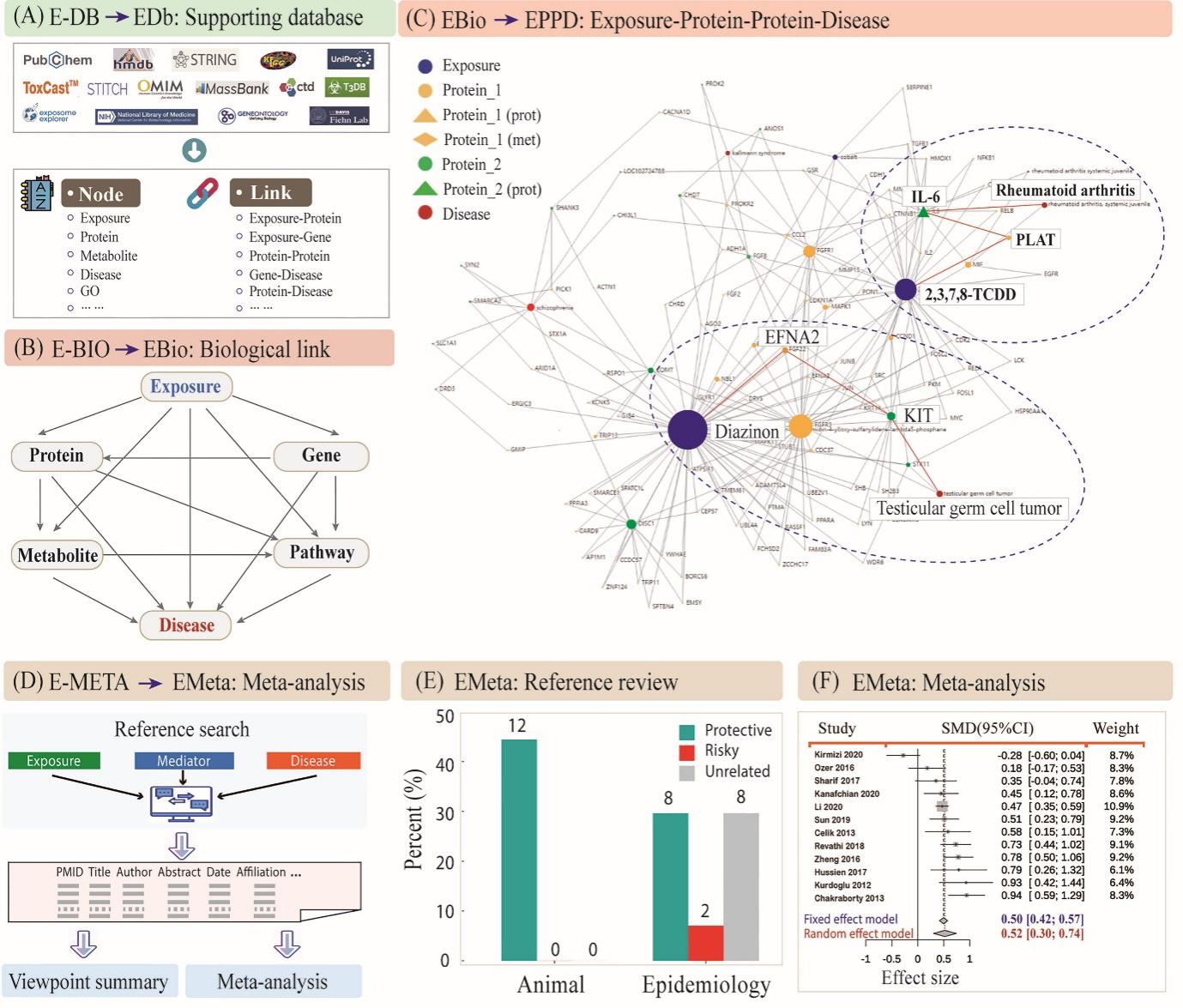
Figure 2. Building biological links between environmental exposure, biological pathway, and health outcome. (A) Framework of the integrated databases of E-BIO function and its searching function; (B) Framework of constructing biological links between environmental exposures and various diseases using EBio module; (C) An application example using the mode of “Exposure-Protein-Protein-Disease” (EPPD) in EBio. The circle size represents the connecting number of the edges for each node. The typical links between exposures and diseases were marked by dashed circles. (D) Framework of building the direct links between environmental exposure and health outcome based on the data-mining technology and the established literature databases (E-META); (E) An application example by reviewing previous publications about the links between zinc intake and asthma risk in EMeta; and (F) An application example by meta-analysis for the previous publications about the association between copper exposure and the risk of polycystic ovary syndrome.
E-MS: Screening Significant Outcome-Related Features from NTA Data
Both positive and negative associations from various epidemiological study designs can be efficiently identified and labeled using the E-MS function (Figure 3A). For example, in a cohort study analyzing the health outcome variable “Y1”, E-MS screened 6,552 features (Figure 3B), and after multiple testing correction, 58 features were found to be significantly associated—demonstrating the function’s capability to prioritize meaningful signals from high-dimensional data. For non-targeted analysis (NTA), E-MS also supports outcome-directed statistical screening by leveraging MS/MS fragmentation data (MS2) to annotate features (Figure 3C), providing a basis for further validation and mechanistic exploration.
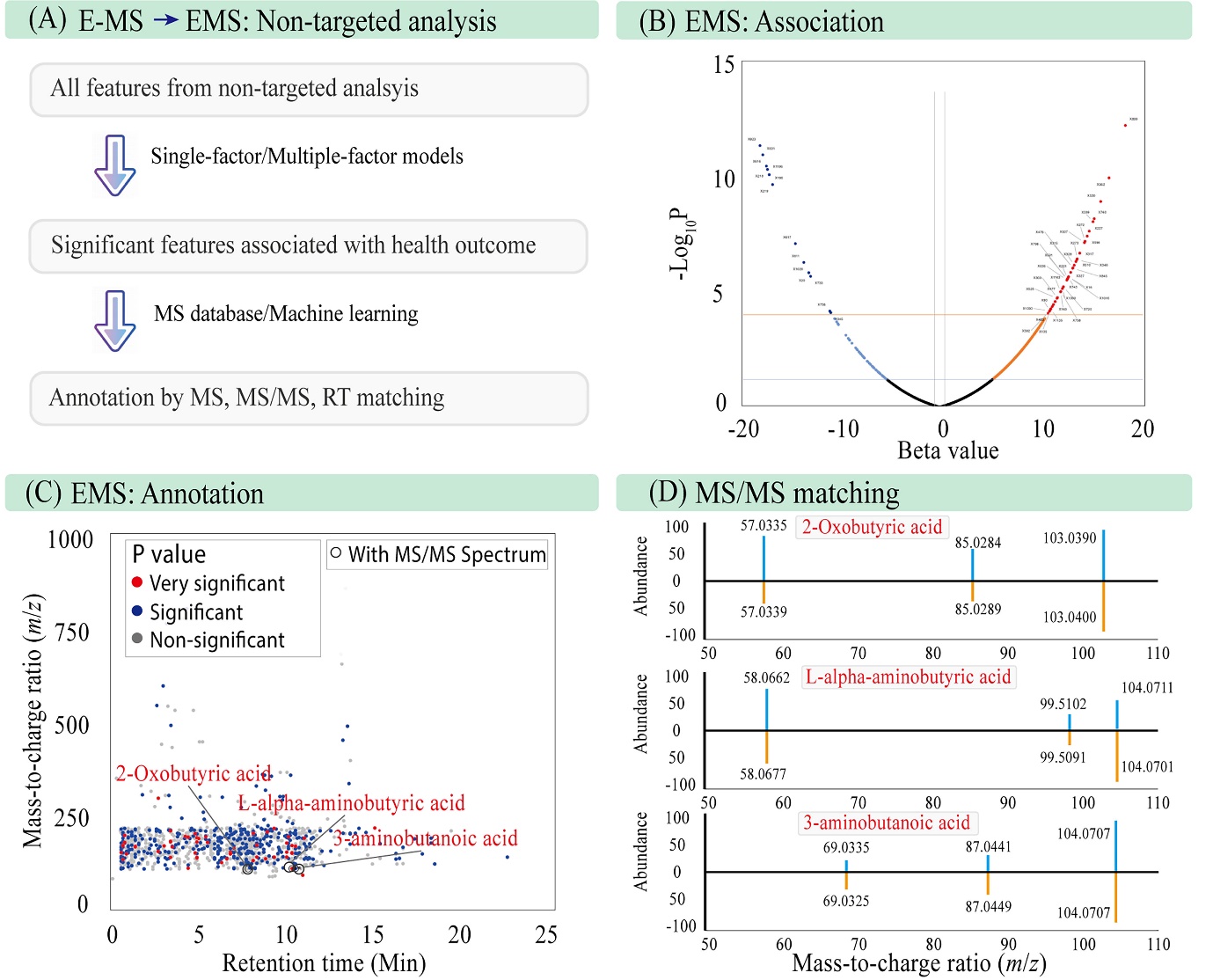
Figure 3. Building E-MS function for the non-targeted analysis using the multi-omics data from MO-Study. (A) Web interface design; (B) Volcano plot of significant features; (C) Annotated features using MS/MS information; and (D) MS/MS matching results for the three chemicals.
E-STAT: Standardization of the Exposomic Analytical Framework and Workflow
The E-STAT module in ExposomeX establishes a standardized framework and operational workflow for exposomic data analysis (Figure 4A). It supports diverse tasks including association (StatCros, StatPanel, StatSurv, StatMedt, StatMix), prediction (StatCros, StatHEP), integration (StatMO), and interpretation (StatIP), tailored to the epidemiological design, data structure, and analytical objectives. The platform accommodates multiple study types—cross-sectional, longitudinal, case-control, cohort, and follow-up—with corresponding data structures such as panel or survival formats. E-STAT incorporates both classical statistical methods (e.g., linear regression, Bayesian models, splines) and machine learning approaches (e.g., decision trees, ensemble learning, stacked generalization). Figure 4B–G illustrates typical modeling results. For instance, in a cohort study, StatCros screened 6,552 features for outcome associations (Figure 4B). Mixture modeling tools like MLR, LASSO, ENET, WQS, G-computation, and BKMR were applied to examine dose–response and interaction effects in mixtures (Figure 4C). To address mediation in complex multivariate settings, an advanced framework integrating environmental risk scores with mediation models was applied, using techniques like pairwise mediation, exposure dimension reduction, and mediator penalization (Figure 4D). Prediction modeling was carried out on single-omic (StatCros, Figure 4E) and multi-omic (StatMO, Figure 4F) data, with models evaluated via ROC curves and integrated using stacked generalization across exposure, metabolome, proteome, and immunome layers. Dimensionality reduction and intra-/inter-omics network construction enhance biological insight. Contribution analysis identified key variables (e.g., X21) driving outcome variation (Figure 4G). Supplementary codes and results are provided with Figure 4.
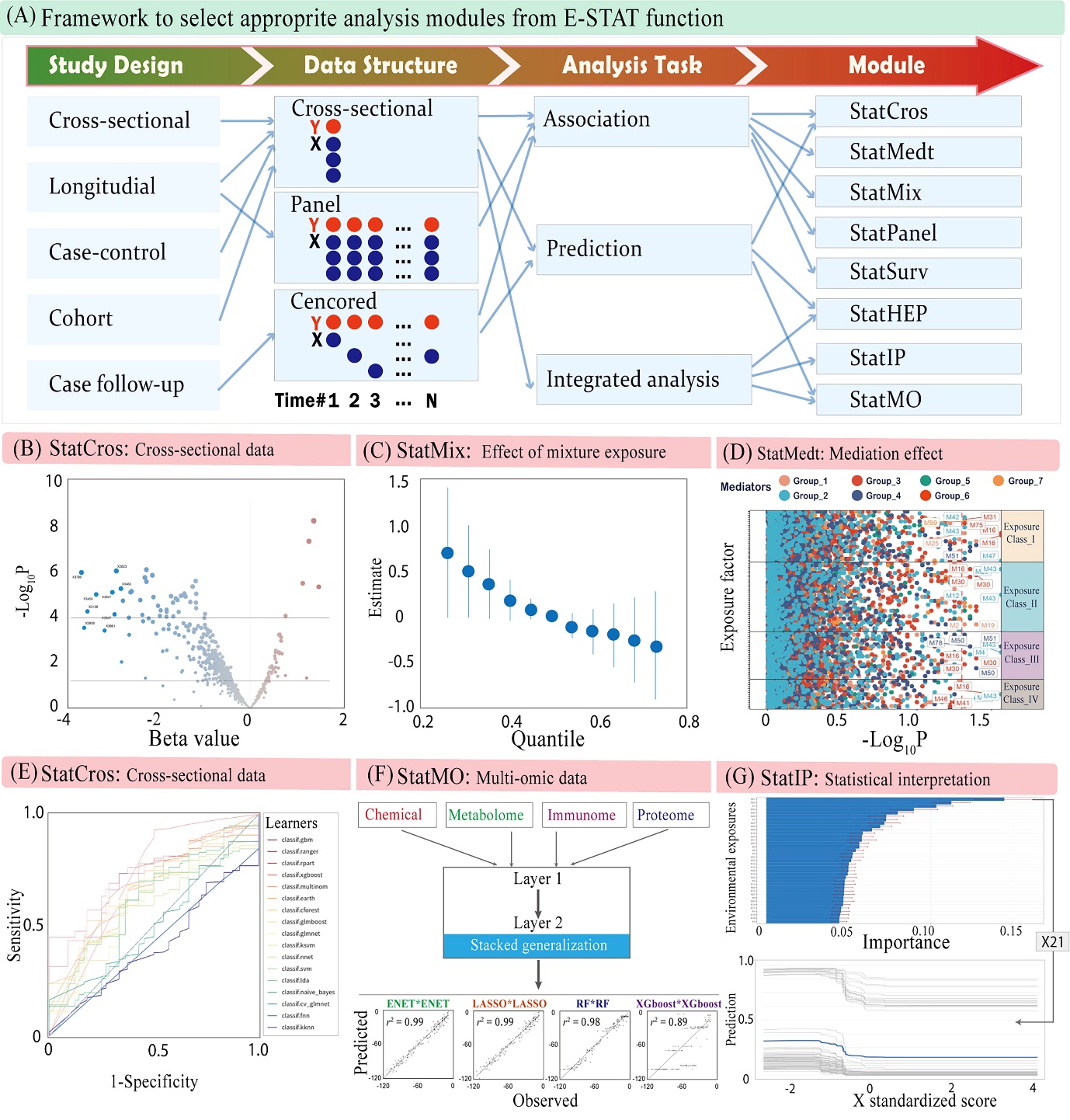
Figure 4. Building statistical models for exposome data. (A) Framework to select appropriate models inconsideration of the epidemiological study design, data structure, and analytical tasks; (B) The effect sizes of association analysis using StatCros. The "Beta value" represents the coefficient of a linear regression model; (C) Exploring the mixture effects of various environmental exposures on health outcomes using StatMix; (D) Screening mediators from the seven potential mediator groups interacting with the four potential exposure groups using StatMedt; (E-F) Prediction analyses for single-omic and multi-omic data using StatCros and StatMO, respectively; and (G) The partial dependence profile of a typical factor (i.e., X21) using StatIP. The variable importance and dose-response trend were calculated using the R package "DALEX" with a random forest algorithm.
E-VIZ: Visualizing the Exploratory Statistical Description
An exploratory descriptive analysis of exposome data can be conducted based on specific epidemiological designs (Figure 5). Before formal analysis, the StatTidy module of E-STAT performs data cleaning, including imputation for missing values and removal of low-variance variables. Data are then transformed through scaling, logarithmic conversion, or dummy encoding, depending on variable type and distribution. Descriptive statistical outputs—such as population summaries ("Table One"), outlier detection, normality testing, group comparisons, and multivariate correlations—are generated using the StatDesc module. For example, in a cohort study with high-dimensional data, population characteristics were easily tabulated (Figure 5A), and further visualized using a heatmap and correlation network, interactively rendered through the E-VIZ module (Figure 5B-D). E-VIZ supports intuitive data exploration through grouped visualization modes: category, relationship, distribution, and component plots (Figure 5E-H).
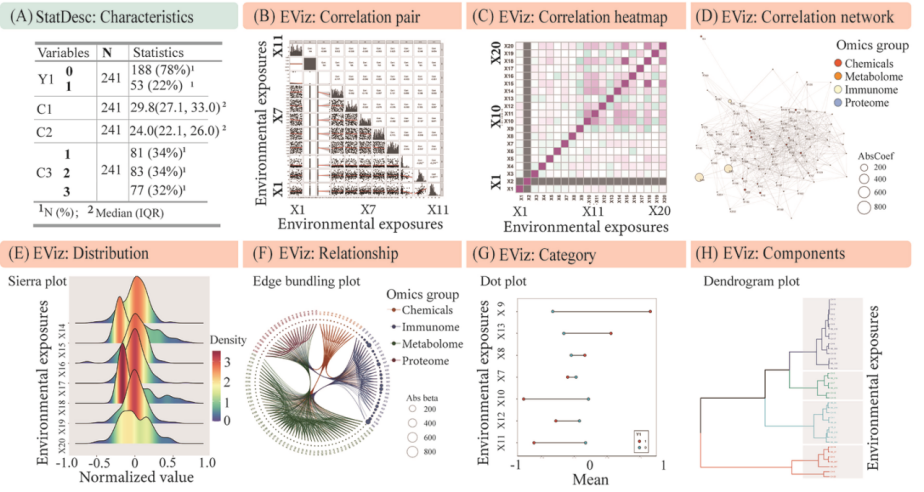
Figure 5. Exploratory analysis by following a specific epidemiological design using E-STAT and E-VIZ functions. (A) Standard table of the demographic information summary in epidemiological studies (i.e. Table one) using StatDesc module; (D) Typical network relationship between features from multiple-omics data using EViz module; (E-H) The typical visualization of the four classes of visualization using EViz including “Distribution” (e.g., sierra plot), “Relationship” (e.g., edge bundling plot), “Category” (e.g., dot plot), and “Components” (e.g., dendrogram plot).
Unique Advantages and Comparative Analysis between ExposomeX and other related platforms
ExposomeX introduces key innovations by integrating curated exposomic data, advanced statistical methods, and high-quality databases into a unified platform that accelerates discovery within the “Exposure–Biology–Disease” framework. Unlike previous platforms, ExposomeX does not merely replicate existing tools but builds a more comprehensive system for exposome analysis. Table 2 compares ExposomeX with platforms like Exposome-Explorer, rexposome, Exposome-NL, HMDB, T3DB, and the Human Blood Exposome DB. Scientifically, ExposomeX establishes the most integrated framework to date, combining statistical modeling with biological insights. It enables construction of exposure–biological–disease networks, going beyond the correlation-focused scope of most existing studies. The platform allows for benchmarking multiple models on resampled datasets, ensuring more robust and unbiased discovery. It also integrates the largest exposome-related database, incorporating resources like CTD, ToxCast, and STITCH, and expands them with data on PM2.5, sleep disorders, and radiation. Modules like E-META and E-MS enhance analytical capacity. In terms of application, ExposomeX is the first platform to offer both an open-source R package and an interactive web interface. Users can access and edit all R scripts and databases—features not commonly supported by platforms like STITCH. The cloud-based web platform enables efficient analysis, sharing, and community contribution. All modules are interoperable, allowing streamlined, end-to-end reproducible workflows.
Table 2. Comparison of Key Features Between ExposomeX and Other Exposome-Related Platforms
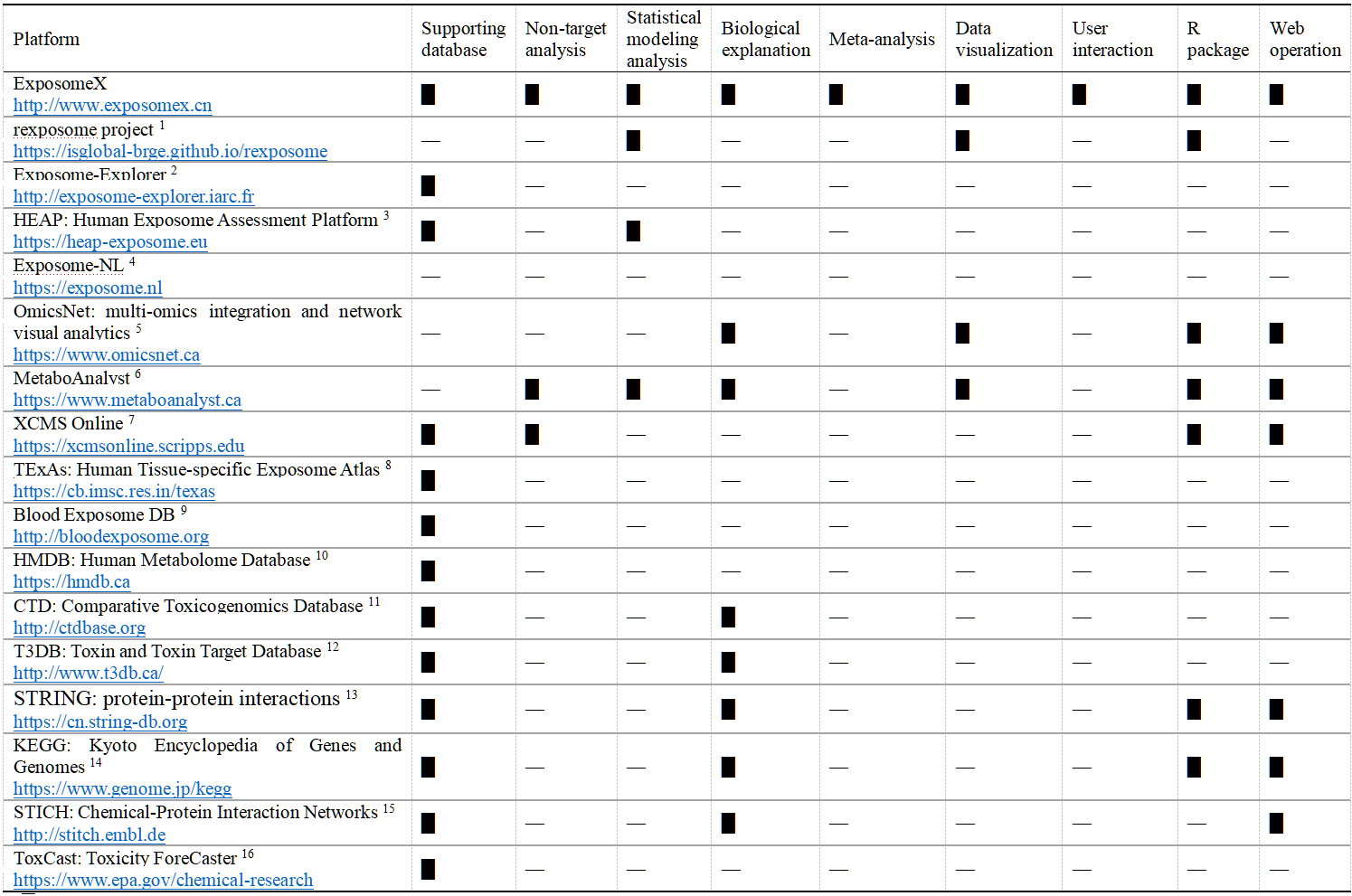
“█” 提供信息; “—”未提供信息
From both scientific and practical standpoints, ExposomeX marks a pioneering advancement in exposome research.
1) Scientific Significance: ExposomeX provides the most integrative analytical framework to date, merging statistical modeling with biological interpretation. It enables the construction and exploration of comprehensive "Exposure–Biology–Disease" networks, advancing beyond traditional multi-omics association studies. The platform supports a wide range of statistical models and allows benchmarking on resampled datasets, enhancing reproducibility and reducing bias. It integrates the largest collection of curated exposome-related databases, including CTD, ToxCast, and STITCH, while incorporating key exposures such as PM2.5, sleep disorders, and radiation. Modules like E-META and E-MS further boost its analytical depth.
2) Application Value: ExposomeX is the first exposome platform to combine an open-source R package with a highly interactive web interface. Users have full access to modify code and databases-capabilities rarely available in other platforms like STITCH. The web interface facilitates cloud-based computation, result sharing, and community contributions. All modules are interoperable, enabling seamless, automated, and reproducible workflows.
Demonstrating its utility, ExposomeX was used to re-analyze three representative studies—mediation analysis (Medt-Study), multi-omics integration (MO-Study), and preterm birth prediction (SPB-Study). Particularly, in SPB-Study, using only 60 samples, ExposomeX effectively applied machine learning to identify key molecular events from complex environmental exposures, showcasing its powerful data-mining capabilities. The platform is officially registered under China’s National Copyright Administration (Figure 6).

Figure 6. Software Copyright Certificate of ExposomeX platform
ExposomeX is an ecosystem platform that integrates R packages with an interactive web interface, built on a unified design philosophy encompassing consistent grammar, data structures, and modular logic. This architecture ensures comprehensive, transparent, traceable, and reproducible exposomic data analysis within an object-oriented computational framework. By standardizing analytical workflows, promoting data sharing, and enabling reproducibility, ExposomeX is poised to make a significant impact on the exposome research field. Its flexible and extensible design allows seamless integration with other tools, increasing adaptability within the scientific community. Future updates will incorporate new statistical methods, expanded data types, and a growing meta-analysis literature database supported by user contributions. Together, these enhancements will strengthen the platform’s capacity to support practical and scalable exposomic research.
We would like to express our gratitude to the working group of environmental exposure and human health of the China Cohort Consortium (http://chinacohort.bjmu.edu.cn). Special thanks to the graduates (Mingyu Li, Baiqiang Li, Yizhong Jin, and Siqi Zhang from North China Electric Power University) for their hard work to develop the web-interface. We also thank Feng Zhao, Siyi Wang, and Jing Yang from Fudan University and Dr. Min Liu, Haoduo Zhao, Junjie Yang and Huili Du from Nanyang Technological University for participating in the work to establish the database and method development, as well as students for code or web testing in Peking University and Fudan University.

